
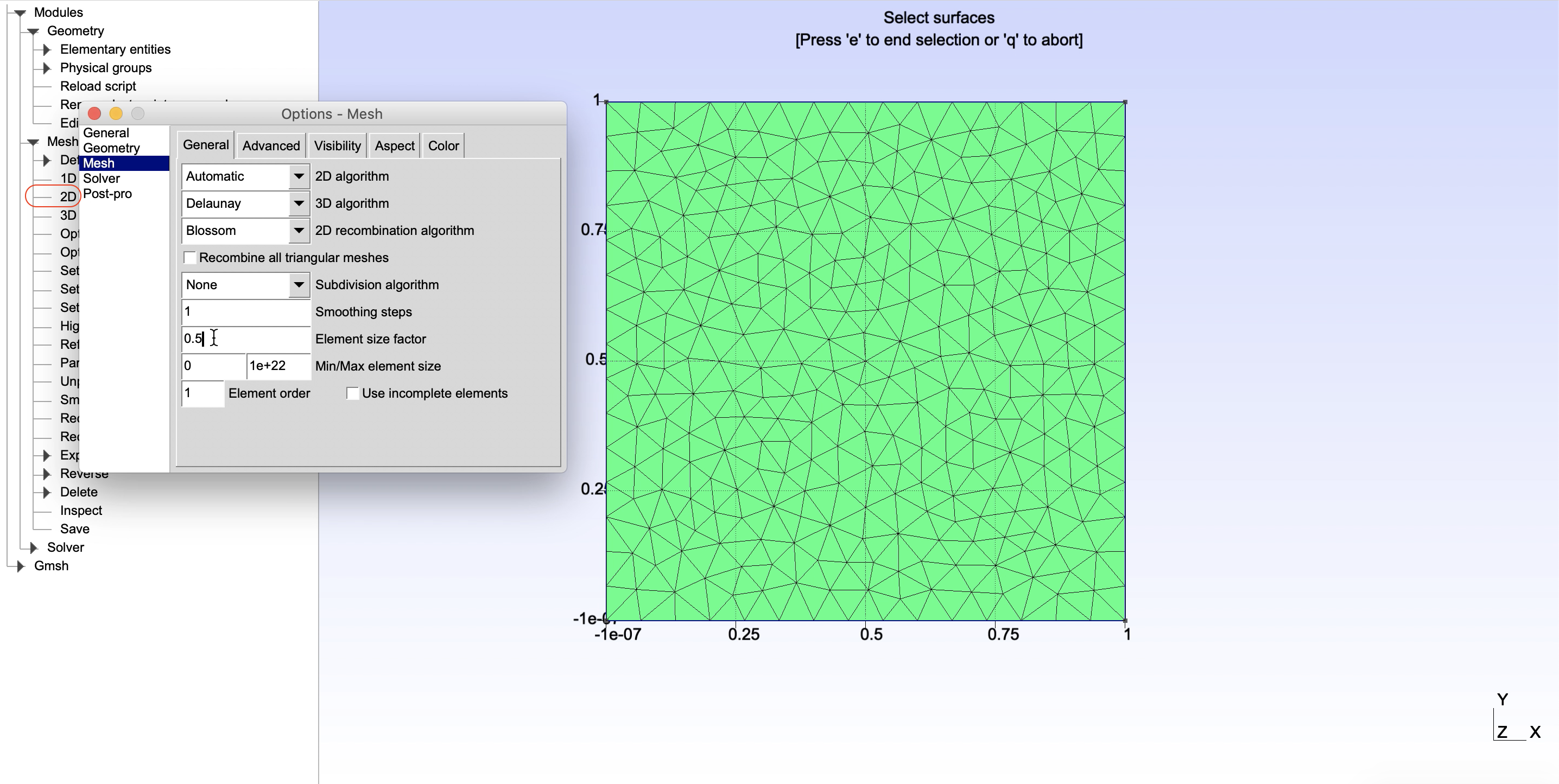
Detailed explanation on the fields can be found on the Gmsh documetation (for example see the explanation t10.geo). Next we will create a structure that will hold the river riv.Geometry = 'Line' īecause we want to refine the mesh around the river we have to provide few additional fields that control the refinment. The direction of the polygon points it is not important, however in case of domains with holes it is very important that the first polygon in the structure is the domain outline. Note that it is necessary to include the nan at the end. Other valid entries that mSim understands are 'Line' and 'Point' dom.X = To generate the mesh we need a structure variable with at least the following fields dom.Geometry = 'Polygon' The shapefiles in Matlab are structure variables with fields. In addition we assume that there are two wells located at (844,1177) and (2356,2829) coordinates and a stream which is described by the line (2500,5000) - (5000,2500). In this example we will create a rectangular domain with size L = 5 km.
#CREATE MESH TUTORIAL GMSH HOW TO#
In this section, we will learn how to load an “msh” file into DOLFINx. In the previous tutorial, we learnt how to load a gmsh python model into DOLFINx. COMM_WORLD, "mesh_out.xdmf", "w" ) as xdmf : xdmf. name = "Facet tags" # Output DOLFINx meshes to fileįrom dolfinx.io import XDMFFile with XDMFFile ( MPI. dim ) adj = AdjacencyList_int32 ( local_entities ) ft = create_meshtags ( mesh, mesh. dim - 1, marked_facets, facet_values ) mesh. int32 ) marked_facets = marked_facets local_entities, local_values = distribute_entity_data ( mesh, mesh. dim - 1, 0 ) gmsh_facet_id = cell_information ] num_facet_nodes = cell_information ] gmsh_facet_perm = perm_gmsh ( facet_type, num_facet_nodes ) marked_facets = numpy. # FIXME: This does not work for prism meshesįacet_type = cell_entity_type ( to_type ( str ( ufl_domain. # Permute facets from MSH to DOLFINx ordering name = "Cell tags" # Create MeshTags for facets dim, 0 ) adj = AdjacencyList_int32 ( local_entities ) ct = create_meshtags ( mesh, mesh. int32 ) local_entities, local_values = distribute_entity_data ( mesh, mesh. We start by specifying a distance field from the obstacle surfaceįrom import AdjacencyList_int32 from import distribute_entity_data from import cell_entity_type, create_meshtags # Create MeshTags for cell dataĬell_values = numpy. One can alternatively set mesh resolutions at points with the command .setSize. The final step is to set mesh resolutions. setPhysicalName ( 2, obstacle_marker, "Obstacle" ) addPhysicalGroup ( 2, obstacles, obstacle_marker ) gmsh.
setPhysicalName ( 2, wall_marker, "Walls" ) gmsh. addPhysicalGroup ( 2, walls, wall_marker ) gmsh. setPhysicalName ( surface, outlet_marker, "Fluid outlet" ) elif np. addPhysicalGroup ( surface, ], outlet_marker ) gmsh. setPhysicalName ( surface, inlet_marker, "Fluid inlet" ) elif np.

addPhysicalGroup ( surface, ], inlet_marker ) inlet = surface gmsh.
getCenterOfMass ( surface, surface ) if np. getEntities ( dim = 2 ) inlet_marker, outlet_marker, wall_marker, obstacle_marker = 1, 3, 5, 7 walls = obstacles = for surface in surfaces : com = gmsh.


 0 kommentar(er)
0 kommentar(er)
